3D-dimensional ultrasound is a very powerful imaging technique, but it requires transducers that accommodate thousands of sensors and complex hardware which restricts its use to specialized clinical applications in developed countries. Manufacturing this type of transducers is complex and very costly. The CUBE multidisciplinary facility and the NeuroComputing Lab (NCL) of the Erasmus Medical Center, Rotterdam, have proposed a novel approach that can overcome this limitation and make 3D imaging simply available to any application by making use of a technique called compressive sensing. The main technological idea behind this technique is that a simple plastic coding mask is already enough to compress the 3D information in such a way that far less sensors are needed. This concept can be applied to extend the imaging capabilities of commercial ultrasound probes from 2D to 3D, but also opens up the possibility for a new type of wearable imaging device for long-time monitoring/imaging of the brain. Key to deploying this technology is the “inverse calculation” of the ultrasound deformations the plastic mask creates while imaging a target (here: neural tissue in living brains). This calculation involves, as a first step, solving a massive linear system of equations, in the range of 30 GB of data per new imaging target. Currency work in the lab involves porting this algorithm onto state-of-the-art GPUs, however we are severely limited by memory-capacity and low-latency concerns, since the system is a streaming, real-time setup.
The goal of this MSc thesis (two independent student topics) is to redeploy the compressive-sensing algorithms onto (a) a Xilinx FPGA, or (b) a Maxeler Dataflow Engine (DFE), for tackling both aforementioned problems. The design should also leave enough silicon space for also extending the algorithms with on-board deep-learning steps, for further image processing. Finally, these accelerated versions should be made to interface to the rest of the experimental setup involving data storage, database access and (offline) image processing. This research is being jointly conducted by the Erasmus Medical Center and the Delft University of Technology.
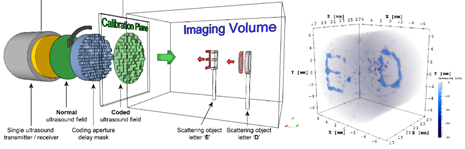
Keywords: HPC, FPGA acceleration, DFE acceleration, image processing, deep learning.
Prerequisites: FPGA design, signal processing (basic), C language, Vivado C language.
Optionally: CUDA programming, image processing, Machine Learning, Deep Learning.
Contact: Christos Strydis
